XP Screens – Intelligent Crystal Screens
XP Screens promote protein crystallization even for most challenging targets and improve diffraction quality of protein crystals. The XP Screens are TEW-optimized JBScreen Basics: 96 of the most prominent crystallization conditions complemented with TEW as “glue” for protein molecules.
There are two variations of the XP Screens:
The XP Screen, SKU M-CS-350, is a convenient initial screen that has successfully induced protein crystallization with low TEW concentrations such as 1 mM. In some cases however, higher concentration of 5 or 10 mM TEW are needed.
The XP Up Screen, SKU M-CS-351, is an upgrade of the well-established XP Screen. It contains 96 of the most prominent screening solutions that are long-term stable in the presence of up to 10 mM TEW.
The Anderson-Evans Polyoxotungstate (TEW), SKU M-X-TEW-5, is a universal and flexible crystallization additive that is integrated the XP Screens. It was shown to improve crystal quality and resolution.
Product Information
The Advantages of XP Screens include:
- Convenient initial screening with up to 10 mM TEW
- Improved crystal quality and resolution
- Tight crystal contacts
- New crystal forms (e.g to provide access to binding pockets)
- Direct phasing
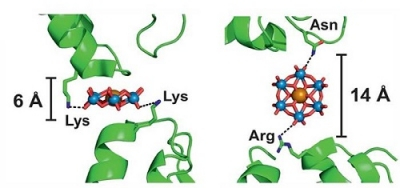
The Anderson-Evans Polyoxotungstate [TeW6 O24] 6- (TEW), SKU M-X-TEW-5, is a universal and flexible crystallization additive that is integrated in the XP Screens. It was shown to improve crystal quality and resolution by:
- Binding to the protein surface and forming tight crystal lattice contacts[3-7]
- Acting as linker in various orientations and thereby creating either smaller
(PDB: 4OUA) or larger (PDB: 4PHI) protein-protein distances[3,4] - Heterogeneous crystallization of two different protein forms in one single crystal
(PDB: 4OUA)[4] - Inducing new crystal forms with access to the active site (PDB: 6ZDK, 6ZDL, 6ZFA)[5]
- Covalently binding and structurally adapting to fit into protein molecules
(PDB: 4Z12, 4Z13)[2]